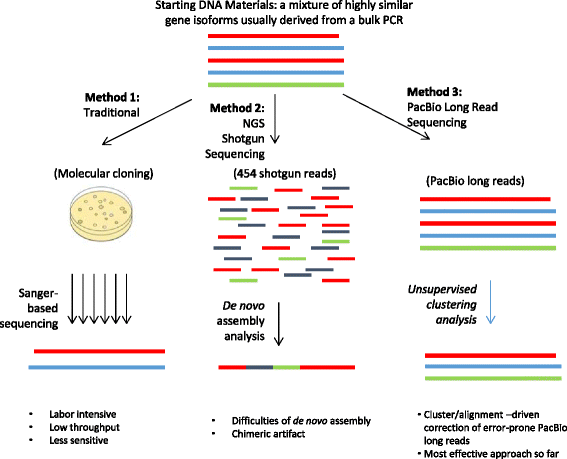
Comparison of three methods for gene isoform identification. Lines
$ 14.00 · 4.6 (198) · In stock

Download scientific diagram | Comparison of three methods for gene isoform identification. Lines with different colors represent different sequences from publication: Distinguishing highly similar gene isoforms with a clustering-based bioinformatics analysis of PacBio single-molecule long reads | Background Gene isoforms are commonly found in both prokaryotes and eukaryotes. Since each isoform may perform a specific function in response to changing environmental conditions, studying the dynamics of gene isoforms is important in understanding biological processes and | PacBio, Bioinformatics Analysis and Nucleic Acid Repetitive Sequences | ResearchGate, the professional network for scientists.

PDF) Distinguishing highly similar gene isoforms with a clustering-based bioinformatics analysis of PacBio single-molecule long reads

Comparison of three methods for gene isoform identification. Lines with

Schema of the clustering-based data analysis procedure using PacBio

Distinguishing highly similar gene isoforms with a clustering-based bioinformatics analysis of PacBio single-molecule long reads, BioData Mining
Distinguishing highly similar gene isoforms with a clustering-based bioinformatics analysis of PacBio single-molecule long reads, BioData Mining

Da Wei (David) HUANG, Big Data Scientist in Bioinformatics, National Institutes of Health, MD, NIH, Bioinformatics and Molecular Analysis Section (BIMAS)

Comparison of three methods for gene isoform identification. Lines with

Brad SHERMAN, Bioinformatics Manager, Master of Science, Leidos Biomedical Research, Inc., Frederick, Laboratory of Human Retrovirology and Immunoinformatics
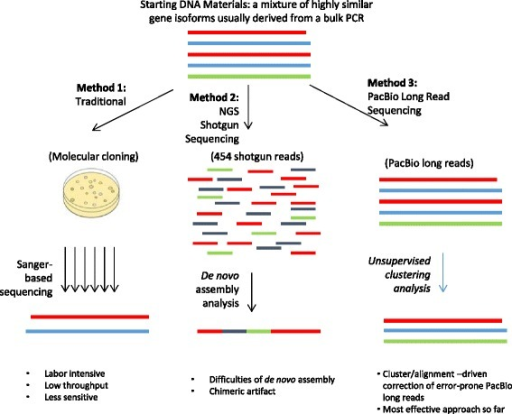
Comparison of three methods for gene isoform identifica

PDF) Distinguishing highly similar gene isoforms with a clustering-based bioinformatics analysis of PacBio single-molecule long reads

Brad SHERMAN, Bioinformatics Manager, Master of Science, Leidos Biomedical Research, Inc., Frederick, Laboratory of Human Retrovirology and Immunoinformatics