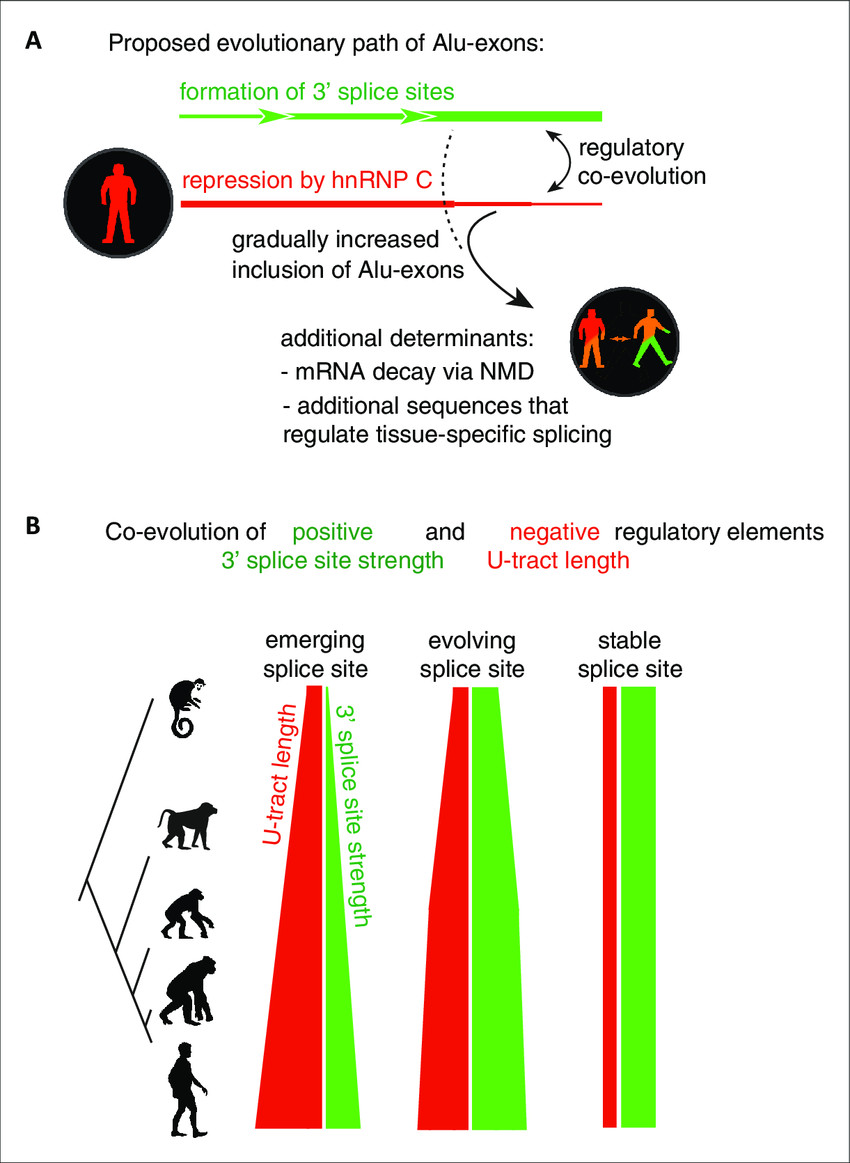
Repressive elements co-evolve with splice site sequences at
$ 25.50 · 4.9 (553) · In stock

Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects

Igor RUIZ DE LOS MOZOS, Computational Research Associate, PhD, The Francis Crick Institute, London, UCL Molecular Neuroscience

Nejc HABERMAN, Imperial College Research Fellow, Doctor of Philosophy, Imperial College London, London, Imperial, Department of Brain Sciences

Repressive elements co-evolve with splice site sequences at cryptic

Evolution of intron–exon structures and alternative splicing of the 3′

U1 snRNP partially rescue the ATT haplotype. (a) Schematic

Igor RUIZ DE LOS MOZOS, Computational Research Associate, PhD, The Francis Crick Institute, London, UCL Molecular Neuroscience
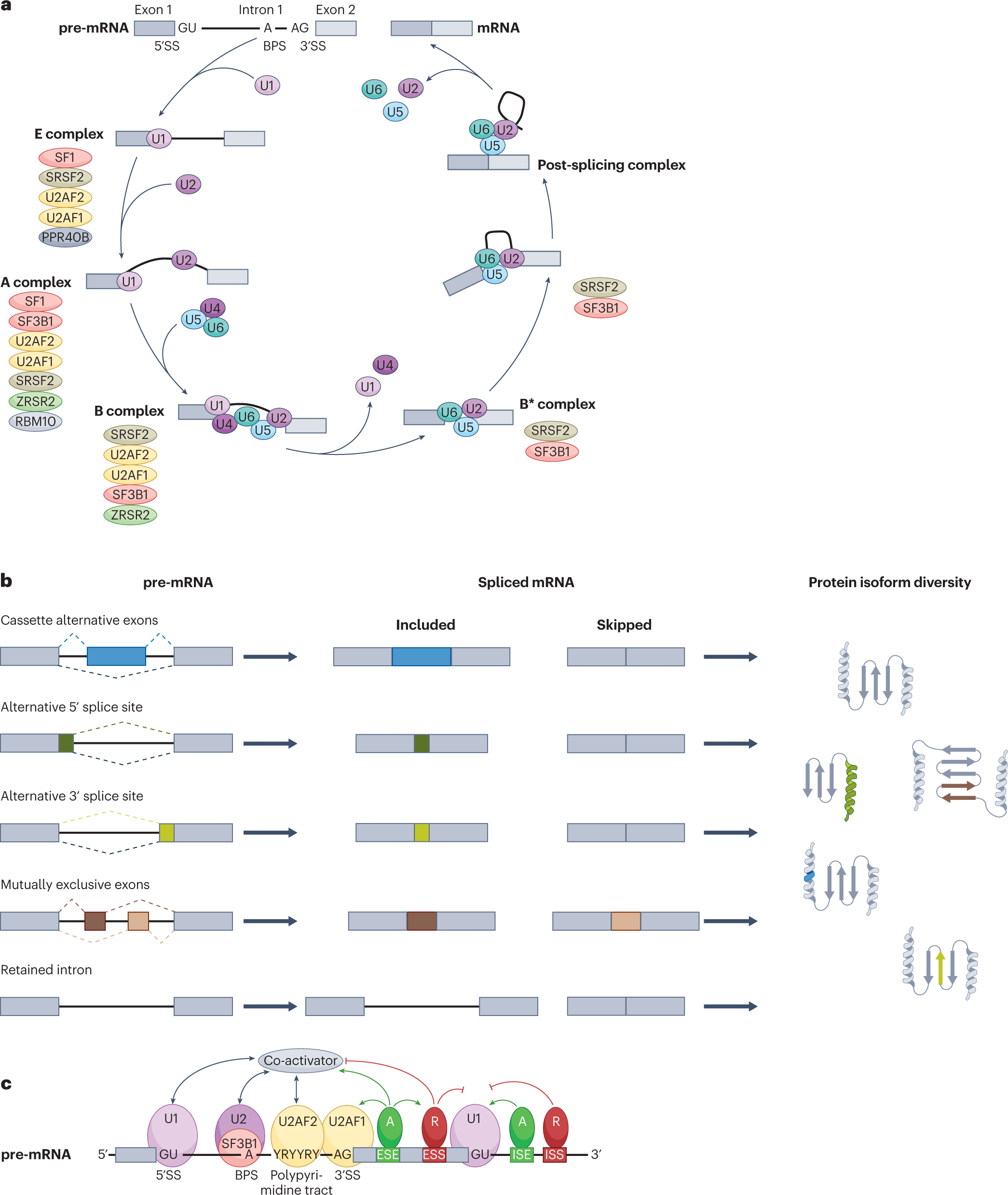
RNA splicing dysregulation and the hallmarks of cancer
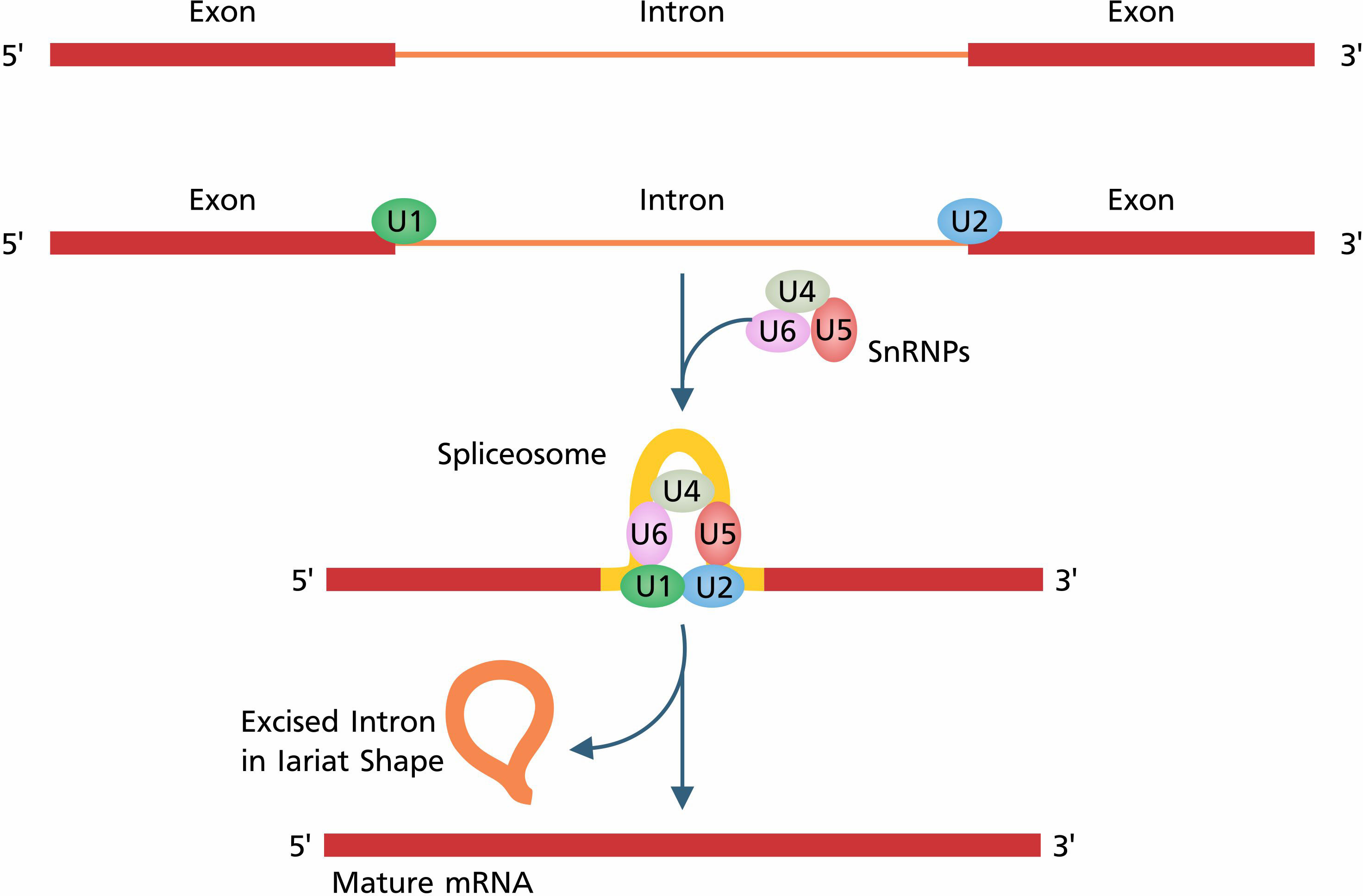
Frontiers Alternative splicing: transcriptional regulatory network in agroforestry

The role of alternative pre-mRNA splicing in cancer progression, Cancer Cell International

Jernej ULE, Professor (Full), PhD, University College London, London, UCL, Institute of Neurology
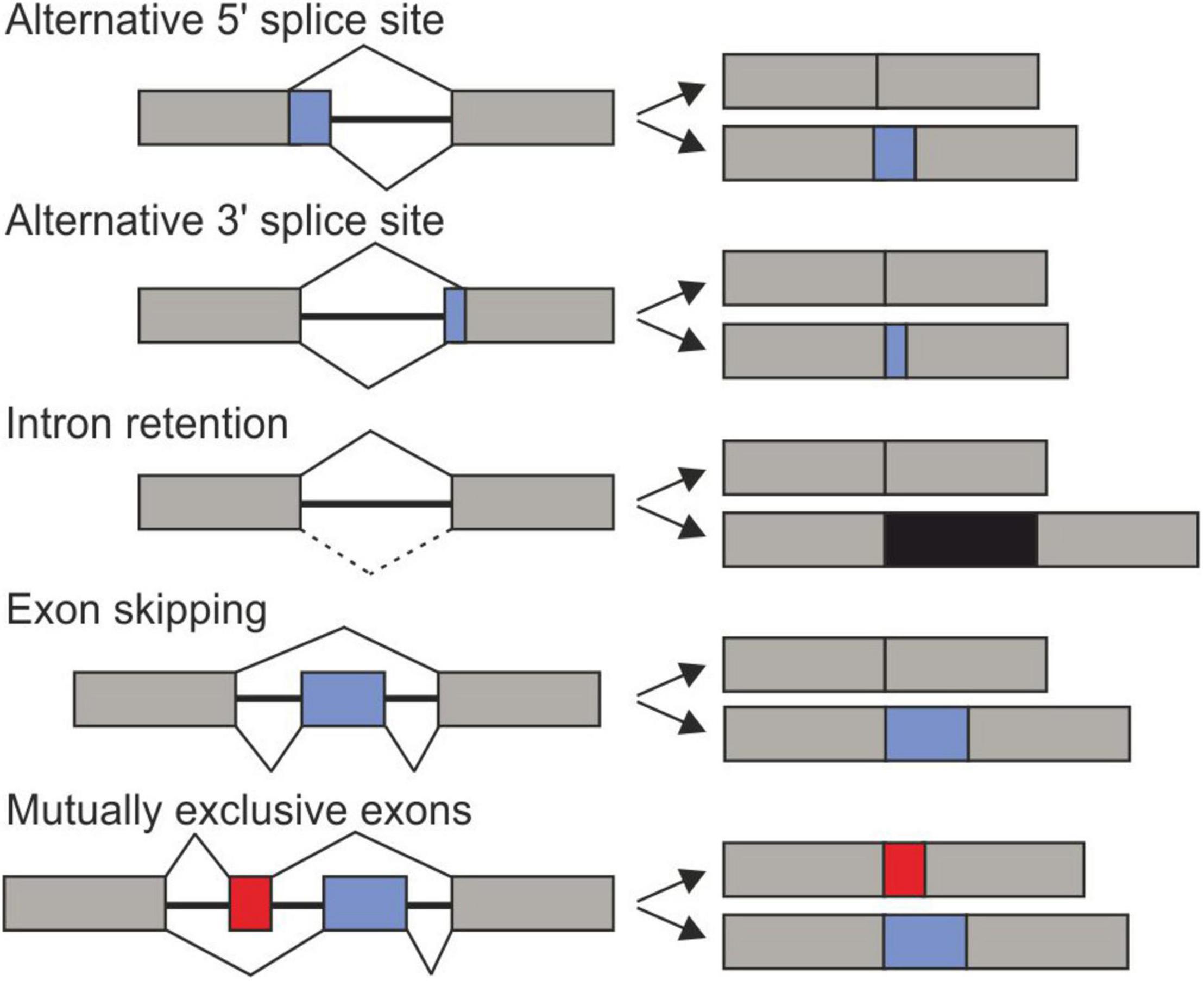
Frontiers Relevance and Regulation of Alternative Splicing in
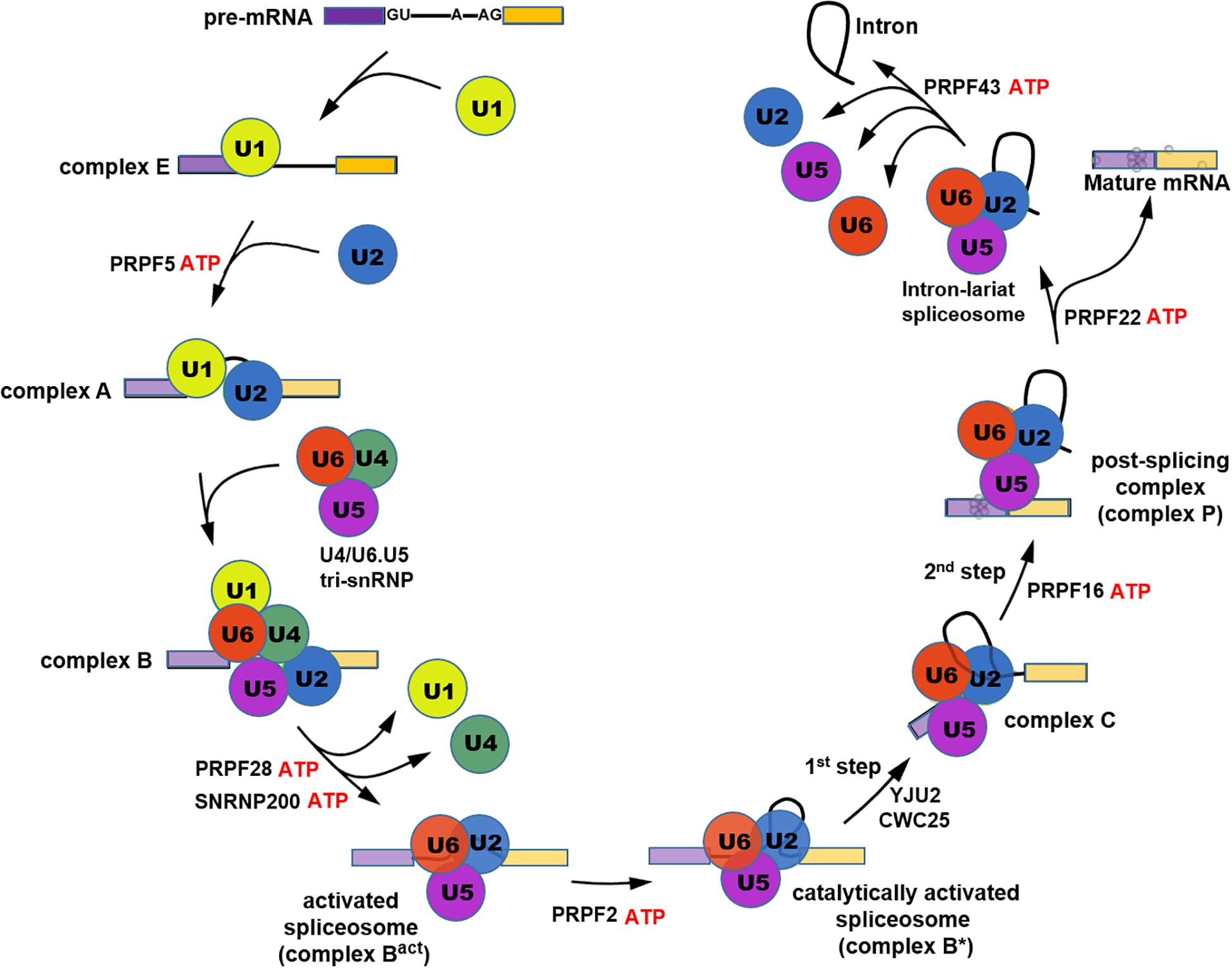
Frontiers Pre-mRNA Processing Factors and Retinitis Pigmentosa: RNA Splicing and Beyond









